
Confirmation bias is a problem that all medical professionals have to wrestle with. Not being able to consider different ideas that challenge pre-existing views blights their ability to entertain a new diagnosis in the face of an established one.
This is a problem rooted in medical literature, where professionals are more likely to lean towards articles that support their pre-existing views. Diversity of opinion is crucial to moulding a holistic perspective capable of delivering effective diagnoses.
In response, a language processing machine learning pipeline was created by this Launchpad App to weed out confirmation bias in medical literature. Using Redis, the Launchpad App created a pipeline that was able to translate text into a knowledge graph through the efficient transmission of data.
Let’s investigate how the team was able to achieve this. But before we dive in, make sure to check out all of the different and innovative apps we have on the Launchpad.
Let’s explore how you can build a pipeline for Natural Language Processing (NLP) using Redis.
We’ll reveal how Redis was used to bring this idea to life by highlighting each of the components used as well as unpacking their functionality.
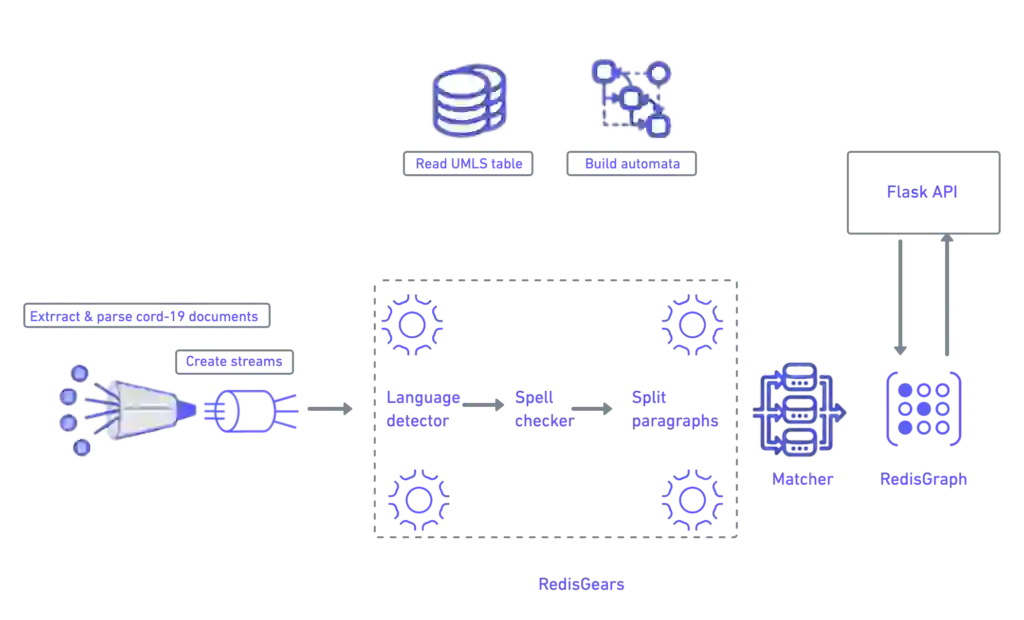
From start to finish, Redis is the data fabric of this pipeline. Its function is to turn text into a knowledge graph. Let’s have a quick overview of what a knowledge graph is and its function in this project.
Systems today go beyond storing folders, files and web pages. Instead, they’re cobwebs of complexity that are composed of entities, such as objects, situations or concepts. A knowledge graph will highlight each of their properties along with the relationship between them. This information is usually stored in a graph database and visualized as a graph structure.
A knowledge graph is made up of 2 main components:
Below is an example of how nodes and edges are used to integrate data.
In the first pipeline, you’ll discover how to create the knowledge graph for medical literature by using a medical dictionary. This processes information using RedisGears and stores it in RedisGraph.
First, use the below code to unzip and parse metadata.zip, where names of files, titles and years are extracted into HASH:
redis_client.hset(f"article_id:{article_id}",mapping={'title': each_line['title']})
redis_client.hset(f"article_id:{article_id}",mapping={'publication_date':each_line['publish_time']})
The following code works by reading JSON files – samples in the data/sample_folder: the-pattern-platform/RedisIntakeRedisClusterSample.py
It also parses JSON into String:
rediscluster_client.set(f”paragraphs:{article_id}“,” “.join(article_body))
And the following code is the main pre-processing task that uses RedisGears: the-pattern-platform/gears_pipeline_sentence_register.py
It also listens to updates on paragraphs: key:
gb = GB('KeysReader')
gb.filter(filter_language)
gb.flatmap(parse_paragraphs)
gb.map(spellcheck_sentences)
gb.foreach(save_sentences)
gb.count()
gb.register('paragraphs:*',keyTypes=['string','hash'], mode="async_local")
This uses RedisGears and HSET/SADD.
How to turn sentences into edges (Sentence) and nodes(Concepts) using the Aho-Corasick algorithm
The first step is to use the following code:
bg = GearsBuilder('KeysReader')
bg.foreach(process_item)
bg.count()
bg.register('sentence:*', mode="async_local",onRegistered=OnRegisteredAutomata)
The next line of code will create a stream on each shard:
'XADD', 'edges_matched_{%s}' % shard_id, '*','source',f'{source_entity_id}','destination',f'{destination_entity_id}','source_name',source_canonical_name,'destination_name',destination_canonical_name,'rank',1,'year',year)
Below is to increase the sentence score:
zincrby(f'edges_scored:{source_entity_id}:{destination_entity_id}',1, sentence_key)
How to populate RedisGraph from RedisGears
the-pattern-platform/edges_to_graph_streamed.py works by creating nodes, edges in RedisGraph, or updating their ranking:
"GRAPH.QUERY", "cord19medical","""MERGE (source: entity { id: '%s', label :'entity', name: '%s'})
ON CREATE SET source.rank=1
ON MATCH SET source.rank=(source.rank+1)
MERGE (destination: entity { id: '%s', label: 'entity', name: '%s' })
ON CREATE SET destination.rank=1
ON MATCH SET destination.rank=(destination.rank+1)
MERGE (source)-[r:related]->(destination)
ON CREATE SET r.rank=1, r.year=%s
ON MATCH SET r.rank=(r.rank+1)
ON CREATE SET r.rank=1
ON MATCH SET r.rank=(r.rank+1)""" % (source_id ,source_name,destination_id,destination_name,year))
the-pattern-api/graphsearch/graph_search.py
Edges with years node ids, limits and years:
"WITH $ids as ids MATCH (e:entity)-[r]->(t:entity) where (e.id in ids) and (r.year in $years) RETURN DISTINCT e.id, t.id, max(r.rank), r.year ORDER BY r.rank DESC LIMIT $limits"""
Nodes:
"""WITH $ids as ids MATCH (e:entity) where (e.id in ids) RETURN DISTINCT e.id,e.name,max(e.rank)"""
Next use the following code to find the most scored articles:
app.py uses zrangebyscore(f"edges_scored:{edges_query}",'-inf','inf',0,5)
Using the most humorous code in the pipeline
Show the below code to your security architect:
import httpimport
with httpimport.remote_repo(['stop_words'], "https://raw.githubusercontent.com/explosion/spaCy/master/spacy/lang/en/"):
import stop_words
from stop_words import STOP_WORDS
with httpimport.remote_repo(['utils'], "https://raw.githubusercontent.com/redis-developer/the-pattern-automata/main/automata/"):
import utils
from utils import loadAutomata, find_matches
This is necessary because RedisGears doesn’t support the submission of projects or modules.
BERT stands for Bidirectional Encoder Representations from Transformers. It was created by researchers at Google AI and is a world leader in processing national language tasks, including Question Answering (SQuAD v1.1.), Natural Language Inference (MNLI) and others.
Below are some popular use cases of the BERT model:
The most advanced code is in the-pattern-api/qasearch/qa_bert.py. This queries the RedisGears + RedisAI cluster, providing users with a question:
get "bertqa{5M5}_PMC140314.xml:{5M5}:44_When air samples collected?"
This queries bertqa prefix on shard {5MP} where PMC140314.xml:{5M5}:44 (see below) is the key of pre-tokenized REDIS AI Tensor (potential answer) and “When air samples collected?” is the question from the user.
PMC140314.xml:{5M5}:44
RedisGears then captures the keymiss event the-pattern-api/qasearch/qa_redisai_gear_map_keymiss_np.py:
gb = GB('KeysReader')
gb.map(qa_cached_keymiss)
gb.register(prefix='bertqa*', commands=['get'], eventTypes=['keymiss'], mode="async_local")
RedisGears then runs the following:
For the non-blocking main thread mode, models are preloaded on each shard using AI.modelset in the-pattern-api/qasearch/export_load_bert.py.
Summarization works by running on sentence: prefix and running t5-base transformers tokenizer, saving results in RedisGraph using simple SET command and python.pickle module, adding summary key (derived from article_id) into:
rconn.sadd('processed_docs_stage3_queue', summary_key)
The following subscribes to queue running simple SET and SREM commands:
summary_processor_t5.py
This will then update hash in RedisGraph:
redis_client.hset(f"article_id:{article_id}",mapping={'summary': output})
Redis provides rgcluster Docker image as well as redis-cluster script. It’s important to make sure that the cluster is deployed in high availability configuration. Each master has to have at least one replica. This is because masters will have to be restarted when you deploy your machine learning models.
If masters is restarted, and there isn’t a replica, then the cluster will become a failed state and you’ll have to manually recreate it.
Other parameters include execution time and memory. Dirt models are 1.4 GB in memory and that’s where you’re required to include proto buffer memory. You also need to increase execution time for both cluster and gears chart because all tasks are computationally extensive, so you need to make sure that the time-outs are accommodated for.
Make sure that you install virtualenv in your system, Docker and Docker compose.
apt install docker-compose
brew install pyenv-virtualenv
git clone --recurse-submodules https://github.com/redis-developer/the-pattern.git
cd the-pattern
./start.sh
cd ./the-pattern-platform/
source ~/venv_cord19/bin/activate #or create new venv
pip install -r requirements.txt
bash cluster_pipeline.sh
Wait for a bit and then check the following:
RedisGraph has been populated:
redis-cli -p 9001 -h 127.0.0.1 GRAPH.QUERY cord19medical "MATCH (n:entity) RETURN count(n) as entity_count"
redis-cli -p 9001 -h 127.0.0.1 GRAPH.QUERY cord19medical "MATCH (e:entity)-[r]->(t:entity) RETURN count(r) as edge_count"
Whether if the API responds
curl -i -H "Content-Type: application/json" -X POST -d '{"search":"How does temperature and humidity affect the transmission of 2019-nCoV"}' http://localhost:8080/gsearch
cd the-pattern-ui
npm install
npm install -g @angular/cli@9
ng serve
cd the-pattern-api/qasearch/
sh start.sh
Note: this will download and pre-load 1.4 GB BERT QA model on each shard. Because of its size, there’s a chance that it might crash on a laptop. Validate by running:
curl -H "Content-Type: application/json" -X POST -d '{"search":"How does temperature and humidity affect the transmission of 2019-nCoV?"}' http://localhost:8080/qasearch
Go to the repository on RedisGears cluster from “the-pattern” repo:
cd the-pattern-bart-summary
# source same enviroment with gears-cli (no other dependencies required)
source ~/venv_cord19/bin/activate
gears-cli run --host 127.0.0.1 --port 30001 tokenizer_gears_for_sum.py --requirements requirements.txt
This task may time out, but you can safely re-run everything.
On the GPU or server, configure NVidia drivers:
sudo apt update
sudo apt install nvidia-340
lspci | grep -i nvidia
sudo apt-get install linux-headers-$(uname -r)
distribution=$(. /etc/os-release;echo $ID$VERSION_ID | sed -e 's/\.//g')
wget https://developer.download.nvidia.com/compute/cuda/repos/$distribution/x86_64/cuda-$distribution.pin
sudo mv cuda-$distribution.pin /etc/apt/preferences.d/cuda-repository-pin-600
sudo apt-key adv --fetch-keys https://developer.download.nvidia.com/compute/cuda/repos/$distribution/x86_64/7fa2af80.pub
echo "deb http://developer.download.nvidia.com/compute/cuda/repos/$distribution/x86_64 /" | sudo tee /etc/apt/sources.list.d/cuda.list
sudo apt-get update
sudo apt-get -y install cuda-drivers
# install anaconda
curl -O https://repo.anaconda.com/archive/Anaconda3-2020.11-Linux-x86_64.sh
sh Anaconda3-2020.11-Linux-x86_64.sh
source ~/.bashrc
conda create -n thepattern_env python=3.8
conda activate thepattern_env
conda install pytorch==1.7.1 torchvision==0.8.2 torchaudio==0.7.2 cudatoolkit=11 -c pytorch
Configure access from instance to RedisGraph docker image (or use Redis Enterprise)
git clone https://raw.githubusercontent.com/redis-developer/the-pattern-bart-summary.git
#start tmux
conda activate thepattern_env
pip3 install -r requirements.txt
python3 summary_processor_t5.py
While RedisGears allows you to deploy and run machine learning libraries like spacy and BERT transformers, the solution below adopts a simpler approach:
Here’s a quick overview of the overall pipeline: The above 7 lines allow you to run logic either in a distributed cluster or on a single machine using all available CPUs. As a side note, no changes are required until you need to scale over more than 1000 nodes.
Use KeysReader registered for namespace paragraphs for all strings or hashes. Your pipeline will need to run in async mode. If you’re a data scientist, we would recommend using gb.run to help ensure that RedisGears works and that it will run in batch mode. It will then run in batch mode. Afterwards, change it to register to capture new data.
By default, functions will return to output, hence the need for count () – to prevent fetching the whole dataset back to the command issuing machine (90 GB for Cord19).
Overall, pre-processing is a straightforward process. You can access the full code here.
You can build Aho-Corasick automata directly from UMLS data. Aho-Corasick will allow you to match incoming sentences into pairs of nodes, and present sentences as edges in a graph. The Gears related code is simple:
bg = GearsBuilder('KeysReader')
bg.foreach(process_item)
bg.count()
bg.register('sentence:*', mode="async_local",onRegistered=OnRegisteredAutomata)
The Redis Knowledge Graph is designed to create knowledge graphs based on long and detailed queries.
Preliminary step: Select ‘Get Started’ and choose either ‘Nurse’ or ‘Medical student.”
Step 1: Type in your query into the search bar
Step 2: Select the query that’s most relevant to your search
Step 3: Browse between the different nodes and change the dates of your search with toggle bar at the bottom
Using Redis, the Launchpad App created a pipeline which helps medical professionals navigate through medical literature without suffering from confirmation bias. The tightly integrated Redis system promoted a seamless transmission of data between components, giving birth to a knowledge graph for medical professionals to utilize.
You can discover more about the ins and outs of this innovative application by visiting the Redis Launchpad. When you’re there, you might also want to browse around our exciting range of applications that we have available.
You can head over to the Launchpad to discover more about the application along with many others in our exciting collections of apps.
Alexander Mikhalev

Alexander is a passionate researcher and developer who’s always ready to dive into new technologies and develop ‘new things.’
Make sure to visit his GitHub page to see all of the latest projects he’s been involved in.